Prostate cancer drug discovery: promising advance

Throughout its collaboration with Johns Hopkins School of Medicine, Kantify identified small molecule candidates by virtually screening millions of entities and testing the expected best ones using a low-throughput screening approach. Only five compounds were tested. This resulted in the identification of four novel entities that could combat prostate cancer.
Prostate cancer
Prostate cancer is a leading cause of death amongst men living in industrialized countries. While early state and localized forms of the disease are nowadays efficiently treated with prostatectomy and radiation therapy, more advanced forms have limited treatment options. Pharmaceutical approaches that are effective in early stages of the disease, which involve lowering androgen levels, often fail at a later stage, when some cancers become resistant to these hormone therapies. Chemotherapy drugs, such as Cabazitaxel, are often considered as a last resort and result in limited improvement in survival rates (see De Bono et al., 2010, for example).
Screening and Collaboration with Johns Hopkins School of Medicine
Perera et al. (2023) previously significantly advanced the field by finding novel biomarkers that clearly discriminate between aggressive and non-aggressive forms of prostate cancer. In order to improve the outcome of patients suffering from these advanced and metastatic forms of prostate cancer, Kantify started a collaboration with Dr. Perera’s lab at Johns Hopkins School of Medicine, with the aim to use Sapian, Kantify's AI drug discovery platform, to discover novel drugs for this indication.
Kantify hypothesized that our target discovery algorithm, Sapian.Target, could identify multiple novel protein targets that drive growth and metastasis in this type of cancer. Five targets in particular scored highly and were shortlisted for the next step. Secondly, we hypothesized that our hit discovery algorithm, Sapian.Hit, could identify polypharmacological new chemical entities (NCE) that acted preferentially on our targets of interest. We virtually screened 5 million compounds and selected three compounds that had a high probability of acting on our targets of interest, and that were chemically dissimilar from each other.
In order to test the effectiveness of the three selected compounds, Dr. Perera’s lab first evaluated the effect of our three candidates on two cancer cell lines, PC3 and LNCaP. These cell lines represent valuable tools in prostate cancer research, offering a simplified yet clinically relevant platform for investigating disease pathogenesis and evaluating potential therapeutic agents.
- PC3 cells, derived from a bone metastasis of prostate adenocarcinoma, exhibit aggressive behavior and androgen receptor independence, making them particularly suitable for studying advanced-stage disease and therapeutic resistance mechanisms.
- LNCAP cells, isolated from a lymph node metastasis, retain androgen receptor sensitivity, reflecting early-stage prostate cancer characteristics and facilitating investigations into hormone-dependent pathways and treatment responses.
Two of the compounds showed that they could significantly inhibit the growth of both cell lines at concentrations below 10 micromolar, as shown by the colony formation assay of Figure 1. As a benchmark, Haynes et al. (2009) conducted a high-throughput screening on approximately 20,000 chemical entities in LNCAP cells, with a resulting hit rate of around 0.5% at a 10 micromolar concentration. Clearly, our workflow resulted in a much higher hit rate (66% - or 132x improvement).
It is also important to note that none of our identified compounds were chemically similar to the hits identified by Haynes, or to any other FDA approved drugs. Our most potent candidate was further validated in vivo through a subcutaneous xenograft tumor assay. The treatment successfully halted tumor growth after a few days, without inducing significant weight loss - a major marker that our compound has a good toxicity profile. The overall selection process is summarized in Figure 2.
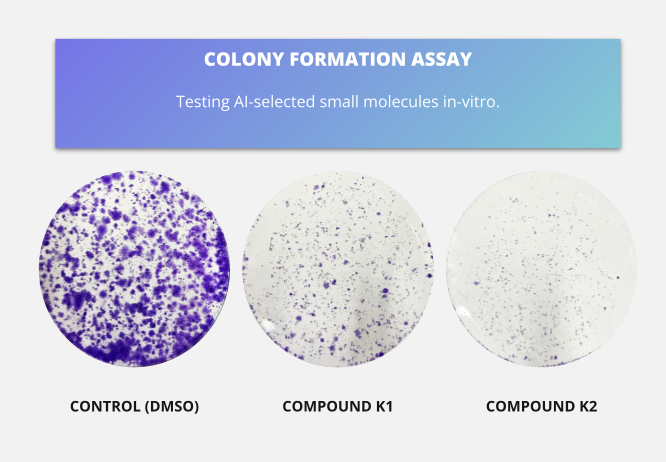
Figure 1: Results from Colony Formation Assay
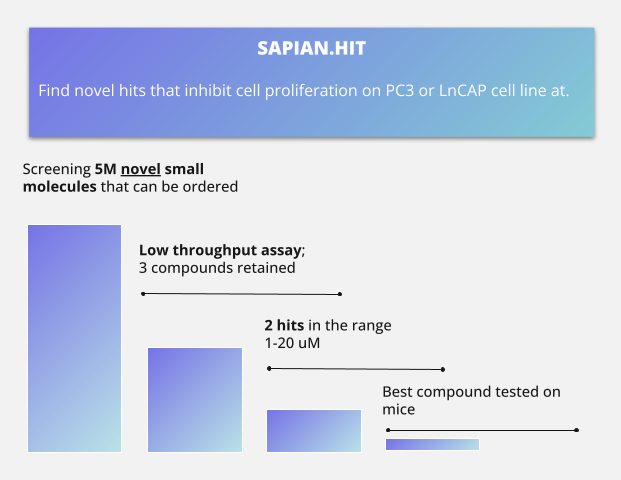
Figure 2: Hit Identification, from Virtual Screening to Running Assays
Finding More Potent Candidates
Building upon the success from the first low throughput screening, Kantify and Dr. Perera worked on identifying additional chemical entities, whose inhibitory effects happen at lower concentrations.
Kantify worked on fine-tuning its core AI model (Sapian.Hit). Two major requirements had to be met from the second batch:
- Small molecules should show efficacy at similar or lower concentrations.
- The inhibition should happen through the same mechanisms of actions as the best hit compound.
Two candidates were retained for the experiment. Both selected compounds exhibited significantly improved potency, reducing the required dose to cause 50% inhibition by a factor of approximately 2. More complete inhibition curves are shown in Figure 3 below.
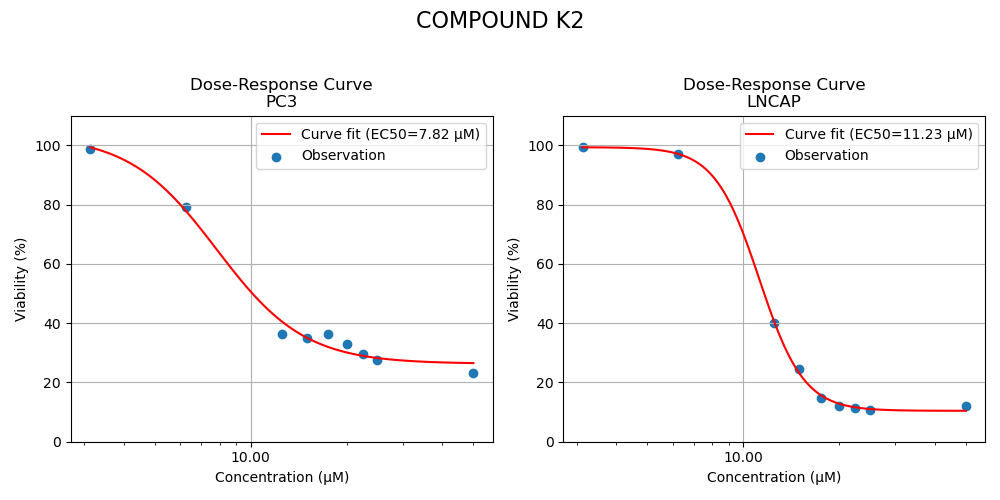
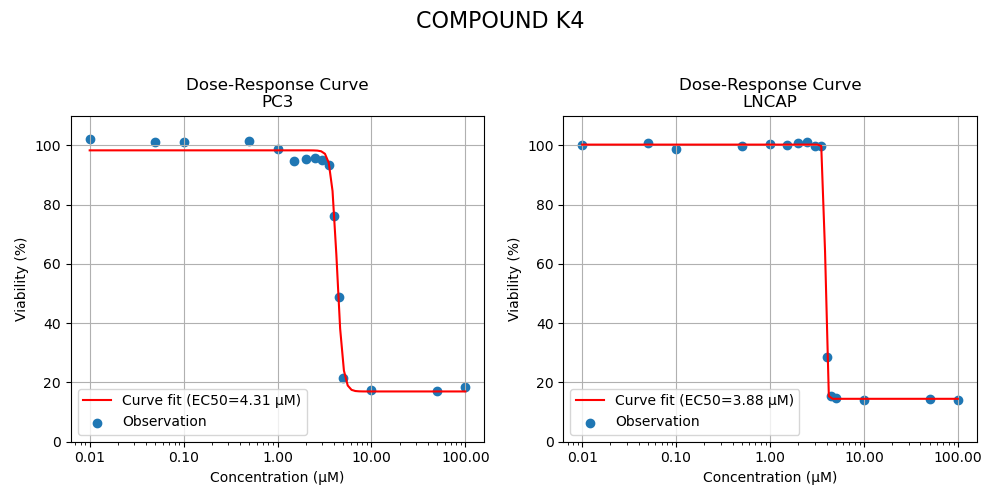
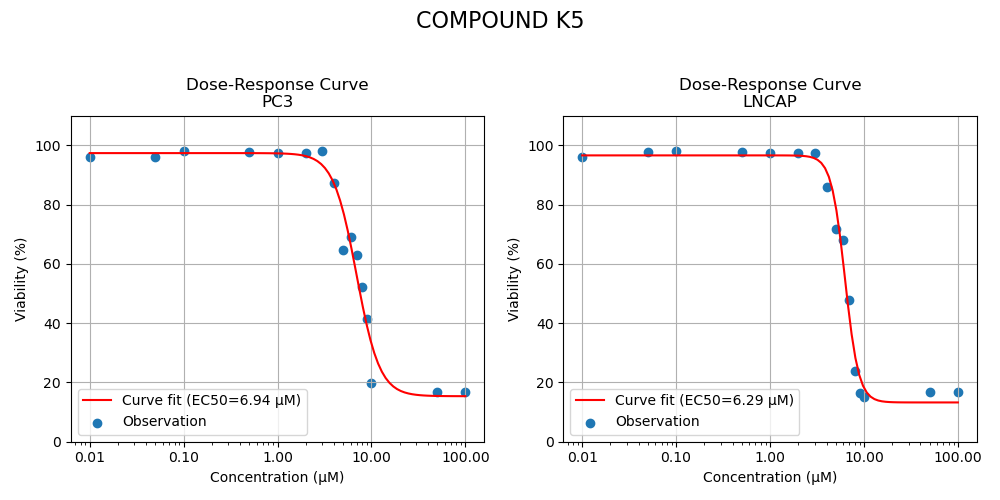
Figure 3: Inhibition curves of compounds K2, K4 and K5
Currently, Dr. Perera and Kantify are further optimizing these compounds to prepare them for human use.
Summary
Throughout its collaboration with Dr. Perera, Kantify identified small molecule candidates by virtually screening millions of entities and testing the expected best ones using a low-throughput screening approach. This resulted in the identification of four novel entities that could combat prostate cancer.
Only five compounds were tested, contrasting with the popular high-throughput screenings, which usually test tens of thousands of compounds. This use of AI in drug screening demonstrates the potential to conduct very low-throughput screening assays on novel chemical entities, which are typically reserved for confirmatory assays. The best hit candidate was found to be safe in mice, a result anticipated by Kantify’s AI platform.
In conclusion, combining AI with the expertise of domain specialists offers a promising pathway to discovering new drugs more quickly, effectively, and cost-efficiently.
Contact us through our contact form below to learn more.